Rodina A, Xu C, Digwal CS, Joshi S, Patel Y, Santhaseela AR, Bay S, Merugu S, Alam A, Yan P, Yang C, Roychowdhury T, Panchal P, Shrestha L, Kang Y, Sharma S, Almodovar J, Corben A, Alpaugh ML, Modi S, Guzman ML, Fei T, Taldone T, Ginsberg SD, Erdjument-Bromage H, Neubert TA, Manova-Todorova K, Tsou MB, Young JC, Wang T, Chiosis G. Systems-level analyses of protein-protein interaction network dysfunctions via epichaperomics identify cancer-specific mechanisms of stress adaptation. Nat Commun. 2023;14(1):3742. doi: 10.1038/s41467-023-39241-7. https://www.nature.com/articles/s41467-023-39241-7
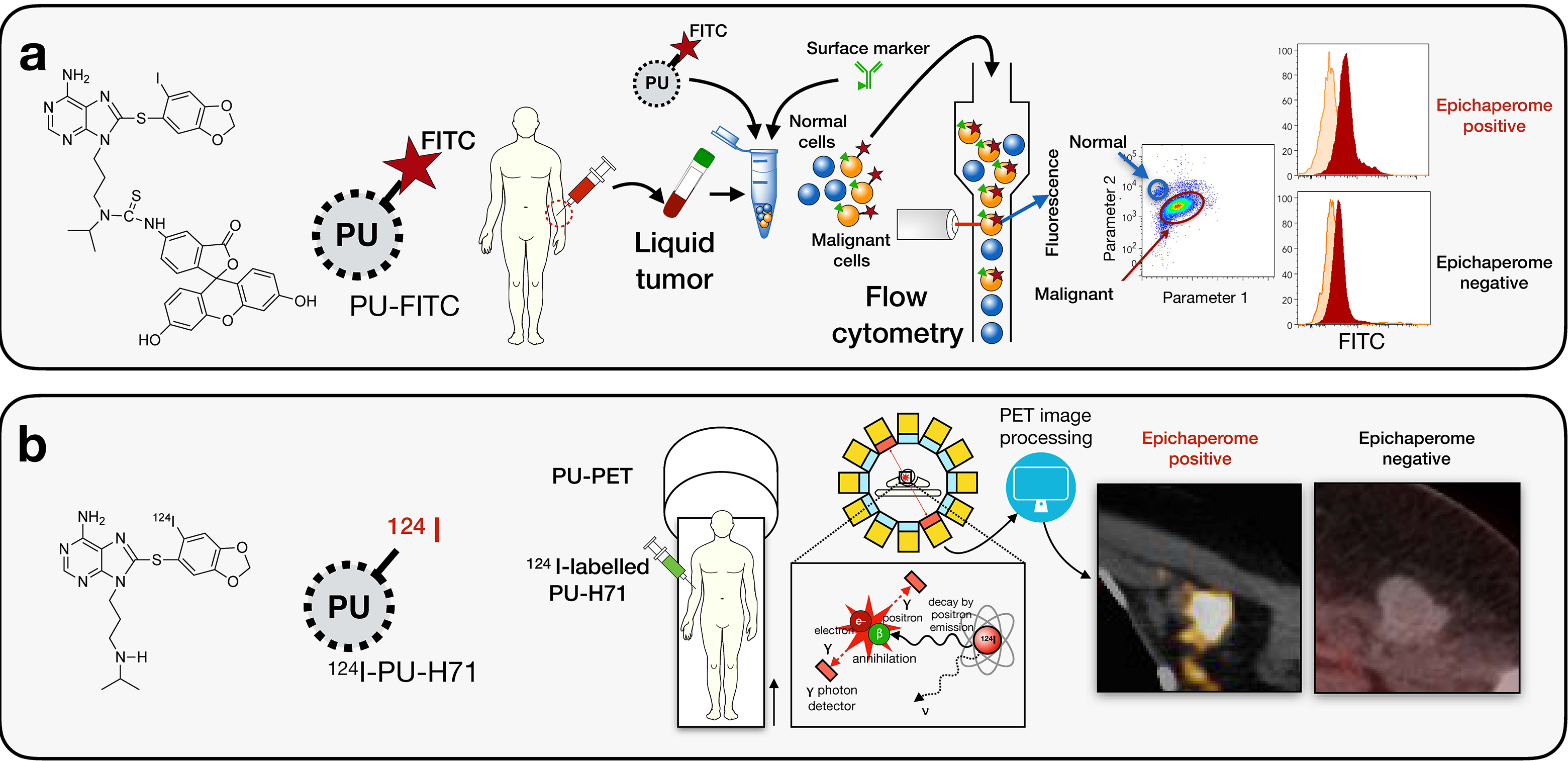
Sharma S, Kalidindi T, Joshi S, Digwal CS, Panchal P, Burnazi E, Lee SG, Pillarsetty N, Chiosis G. Synthesis of (124)I-labeled epichaperome probes and assessment in visualizing pathologic protein-protein interaction networks in tumor bearing mice. STAR Protoc. 2022;3(2):101318. doi: 10.1016/j.xpro.2022.101318. https://pubmed.ncbi.nlm.nih.gov/35496791/
Alam A, Wang T, Chiosis G. Cytoscape files—Systems-level analyses of protein-protein interaction network dysfunctions via epichaperomics identify cancer-specific mechanisms of stress adaptation [Data set]. Zenodo https://doi.org/10.5281/zenodo.7433980 (2022). https://zenodo.org/record/7433980
Wang T, Digwal CS, Alam A, Chiosis G. R Script Epichaperomics—Systems-level analyses of protein-protein interaction network dysfunctions via epichaperomics identify cancer-specific mechanisms of stress adaptation. Zenodo https://doi.org/10.5281/zenodo.7416220 (2022). https://zenodo.org/record/7416220
Ginsberg SD, Neubert TA, Sharma S, Digwal CS, Yan P, Timbus C, Wang T, Chiosis G. Disease-specific interactome alterations via epichaperomics: the case for Alzheimer’s disease. FEBS J. 2021 May 24. doi: 10.1111/febs.16031. Epub ahead of print.
Sugita M, Wilkes DC, Bareja R, Eng KW, Nataraj S, Jimenez-Flores RA, Yan L, De Leon JP, Croyle JA, Kaner J, Merugu S, Sharma S, MacDonald TY, Noorzad Z, Panchal P, Pancirer D, Cheng S, Xiang JZ, Olson L, Van Besien K, Rickman DS, Mathew S, Tam W, Rubin MA, Beltran H, Sboner A, Hassane DC, Chiosis G, Elemento O, Roboz GJ, Mosquera JM, Guzman ML. Targeting the epichaperome as an effective precision medicine approach in a novel PML-SYK fusion acute myeloid leukemia. NPJ Precis Oncol. 2021 May 26;5(1):44. doi: 10.1038/s41698-021-00183-2.
Jhaveri KL, Dos Anjos CH, Taldone T, Wang R, Comen E, Fornier M, Bromberg JF, Ma W, Patil S, Rodina A, Pillarsetty N, Duggan S, Khoshi S, Kadija N, Chiosis G, Dunphy MP, Modi S. Measuring Tumor Epichaperome Expression Using [124I] PU-H71 Positron Emission Tomography as a Biomarker of Response for PU-H71 Plus Nab-Paclitaxel in HER2-Negative Metastatic Breast Cancer. JCO Precis Oncol. 2020 Nov 17;4:PO.20.00273. doi: 10.1200/PO.20.00273.
Pillarsetty N, Jhaveri K, Taldone T, Caldas-Lopes E, Punzalan B, Joshi S, Bolaender A, Uddin MM, Rodina A, Yan P, Ku A, Ku T, Shah SK, Lyashchenko S, Burnazi E, Wang T, Lecomte N, Janjigian Y, Younes A, Batlevi CW, Guzman ML, Roboz GJ, Koziorowski J, Zanzonico P, Alpaugh ML, Corben A, Modi S, Norton L, Larson SM, Lewis JS, Chiosis G, Gerecitano JF, Dunphy MPS. Paradigms for Precision Medicine in Epichaperome Cancer Therapy. Cancer Cell. 2019 Nov 11;36(5):559-573.e7. doi: 10.1016/j.ccell.2019.09.007. Epub 2019 Oct 24.
Dunphy MPS, Pressl C, Pillarsetty N, Grkovski M, Modi S, Jhaveri K, Norton L, Beattie BJ, Zanzonico PB, Zatorska D, Taldone T, Ochiana SO, Uddin MM, Burnazi EM, Lyashchenko SK, Hudis CA, Bromberg J, Schöder HM, Fox JJ, Zhang H, Chiosis G, Lewis JS, Larson SM. First-in-Human Trial of Epichaperome-Targeted PET in Patients with Cancer. Clin Cancer Res. 2020 Oct 1;26(19):5178-5187. doi: 10.1158/1078-0432.CCR-19-3704. Epub 2020 May 4.