Our lab has had a long-standing interest in the role and regulation of the p53 tumor suppressor. Mutations in the p53 gene (TP53) are often associated with aggressive tumor behavior and poor patient prognosis, and p53 mutant tumors display rampant genomic instability (1). Wild-type p53 is often engaged in response to cellular damage, where it acts to regulate genes that promote cell-cycle arrest, apoptosis, senescence, differentiation, and autophagy (Figure 1). In addition to these cell-autonomous activities, p53 can promote the secretion of a variety of factors that influence the tissue microenvironment. A major unanswered question in the field is what p53 effector functions mediate its tumor suppressor functions and how their disruption gives rise to highly aggressive tumors.
Historically our laboratory has emphasized understanding how p53 regulates apoptosis and cellular senescence, though we have also become interested in the ability of p53 to restrict lineage plasticity (2). Using a murine model of pancreatic ductal adenocarcinoma (PDAC) in which endogenous p53 can be switched on and off using a tetracycline-inducible shRNA, we showed that restoring p53 in advanced PDAC increased a-ketoglutarate levels and activated a transcriptional program that drove tumor cells into a premalignant state (3). These observations imply that p53 loss is required to sustain tumor cells in a less-differentiated state and illustrate how mimicking p53 function might have therapeutic benefit. Current efforts combine a range of multiomic and spatial imaging approaches to understand p53 action and the immediate effects of p53 loss in sporadic cancer progression.
p53 inactivation typically arises through a missense mutation in one allele and a “second hit” deletion event that also encompasses adjacent genes (1). Researchers often regard all p53 mutations as equivalent, though some missense mutants apparently have “gain-of-function” effects (4). Moreover, the p53 deletions frequently encompass many other genes. Using chromosome engineering, we also showed that the deletion events target biologically relevant genes whose co-deletion enhances tumorigenesis (5). These results challenge the concept of classifying tumors simply as p53 wild-type or mutant. Current efforts in the laboratory are using new CRISPR base-editing methodologies to better understand the functional consequences of allelic variation in p53 mutations.
Addressing the role of “genomic instability” in the evolution of p53 mutant tumors, we used a unique model that enables fluorescent tagging of cells that inactivate p53 well before tumors arise to characterize genomic changes from premalignancy to full-blown cancer (6). The work demonstrated a deterministic pattern of genome evolution involving copy-number alterations that contribute to the malignant potential of p53-mutant cells (Figure 2). Thus, despite the chaotic nature of p53-mutant tumors, their evolution follows rules that may eventually be exploited. Current efforts in the laboratory focus on other aspects of how cells that acquire p53 mutations evolve towards cancer.
Complex Karyotype Acute Myeloid Leukemia as a Paradigm of a p53 Mutant Cancer
Complex karyotype acute myeloid leukemia (CK AML) is a lethal cancer that frequently harbors p53 mutations. Unlike most other myeloid leukemias, but consistent with the action of p53 dysfunction mentioned on our p53 tumor suppressor network page, CK AML is defined by the presence of three or more detectable chromosomal abnormalities that often occur at recurrent regions of the genome. CK AML is resistant to standard chemotherapy, and patients have few good treatment options. Building on efforts in other leukemia subtypes (7, 8), we are investigating how p53 restricts leukemia development and how p53 mutations cooperate with copy number alterations to give rise to aggressive disease (9, 10). Our goal is to identify strategies to therapeutically target CK AML.
Recent efforts to understand how p53 mutant status in CK AML drives disease has identified haploinsufficient tumor suppressor genes linked to p53 that are important in AML development and a gain-of-function activity for one p53 mutant in AML (4, 5). We are currently expanding our efforts to characterize other p53 mutant alleles and identifying molecular vulnerabilities unique to CK AML.
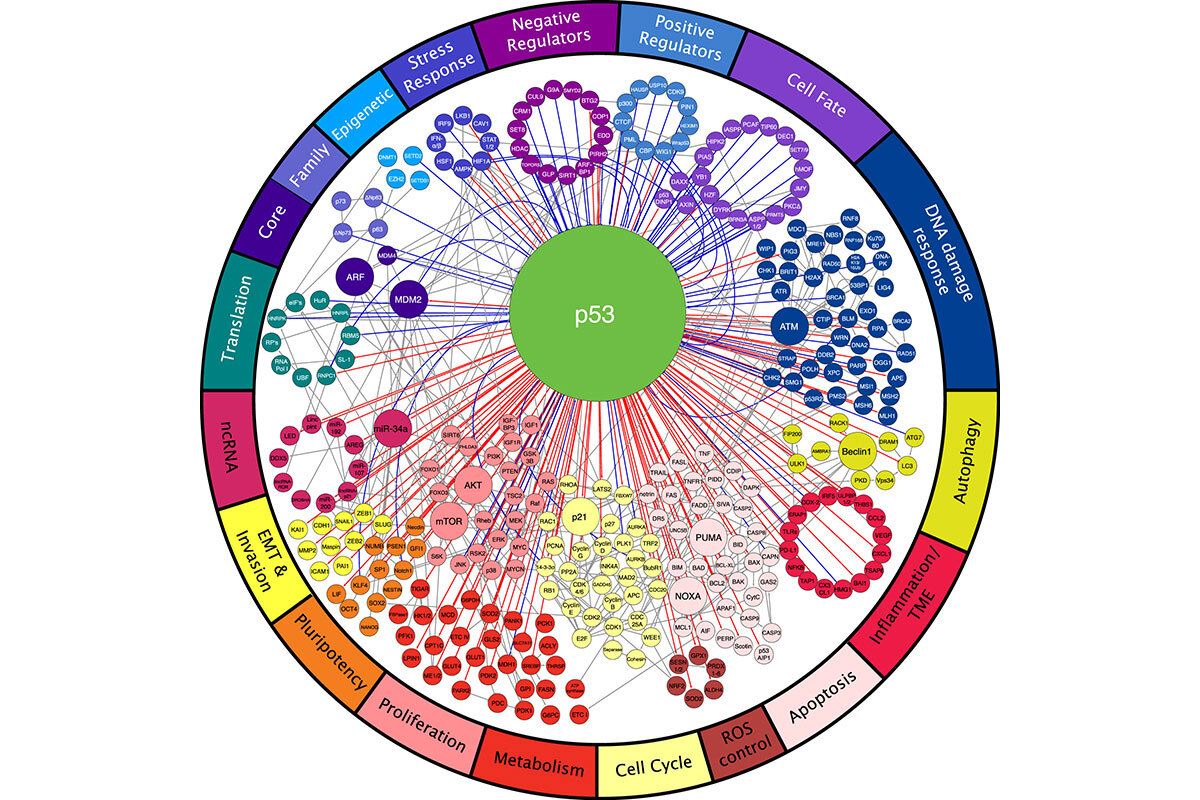
Figure 1. p53 acts within a network of proteins to regulate a broad array of biological processes (18).
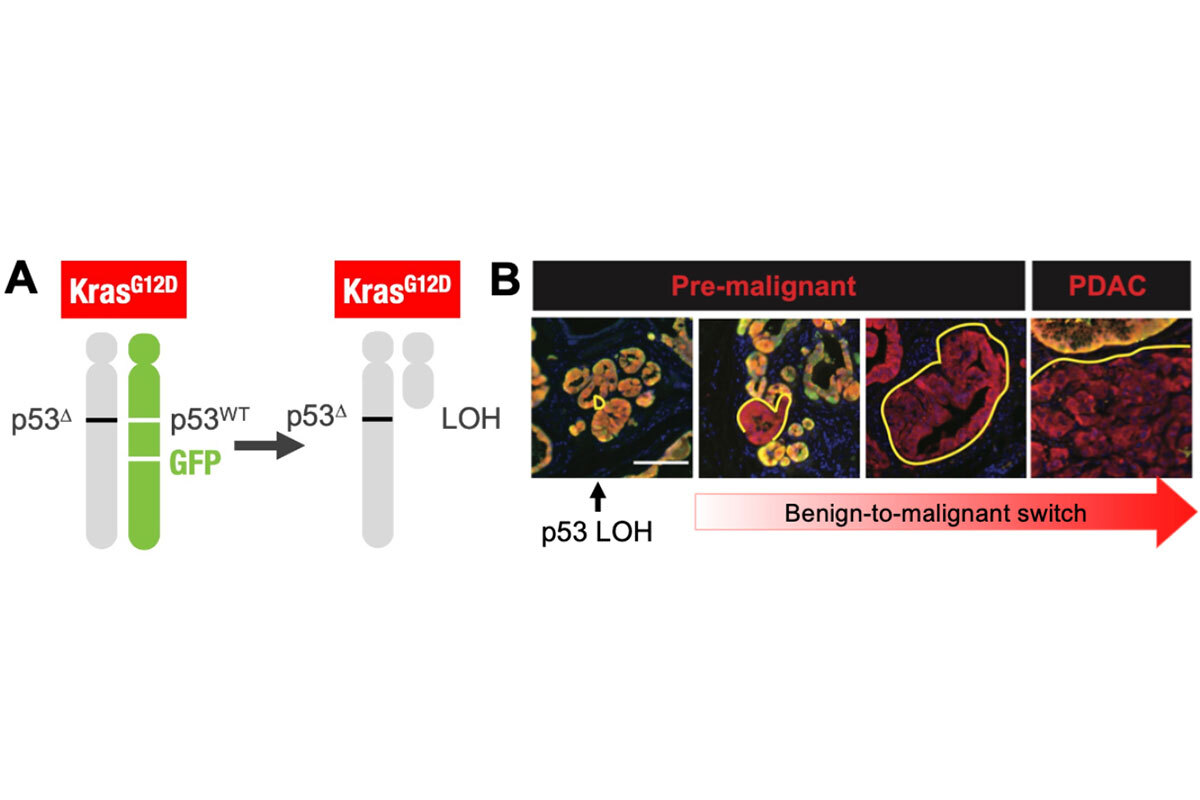
Figure 2. Model for tracking loss of p53 in developing tumors. The model is engineered such that pancreatic epithelial cells become KrasG12D, p53-/+ and express mKate (red) and GFP when mice are given doxycycline. Because the GFP cassette is linked to the wild-type p53 allele, individual cells that then undergo loss of heterozygosity to lose the wild-type p53 allele become mKate+/GFP- and can be tracked and isolated throughout tumor evolution.