Age-old epidemiologic studies indicate that cancer is influenced by environmental factors. While some factors act by altering mutation rates, others work through epigenetic mechanisms that we suspect blunt tumor-suppressive programs and/or alter cell state. Our initial foray into cancer epigenetics involved efforts to understand how mutations in chromatin remodeling factors promote cancer (1, 2) and through functional genomics screens to identify cancer vulnerabilities (3, 4).
In a new line of research, we have begun to study how epigenetic programs contribute to gene–environment interactions in cancer. As one paradigm (5), we explored how tissue damage/inflammation accelerates Kras-driven early neoplasia in the pancreas. With Dana Pe’er, we defined the specific epigenetic programs that distinguish early neoplasia from normal regeneration and characterized cell state and cell-cell communication changes unique to damage-induced neoplasia (Figure 1) (6, 7). These studies show that inflammation and Kras mutations cooperate to stimulate the emergence of cells with cancer-like epigenetic states within 48 hours of tissue damage. We continue to study how inflammation and other environmental factors influence the output of cancer-promoting gene mutations. Understanding these processes may reveal new vulnerabilities not predicted through genetics alone.
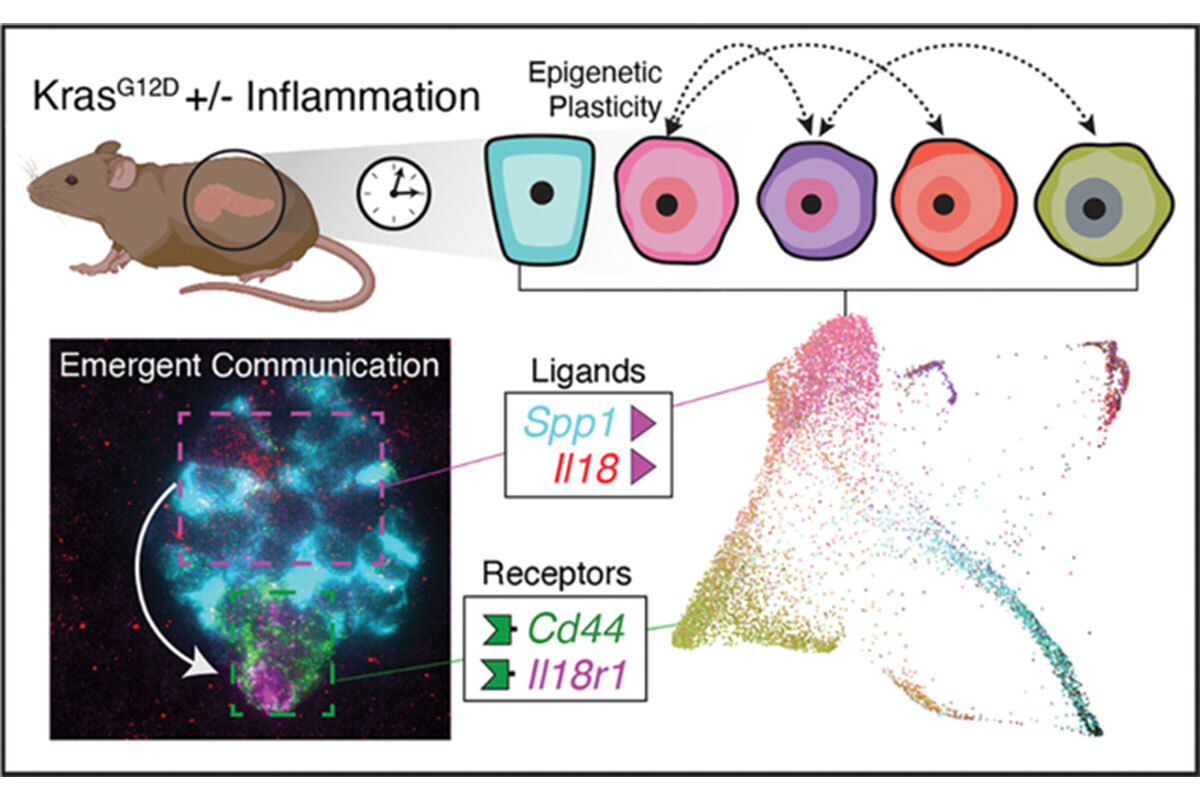
Figure 1. Cell states and cell-cell communication in early pancreatic tumorigenesis inferred from multiplexed single-molecule fluorescence in situ hybridization (lower left) and single-cell RNA sequence data (lower right) (57). Mouse illustration was created with BioRender (www.biorender.com).