DNA replication, repair, and damage response, collectively called the 3Rs, support genome stability and faithful genetic inheritance. Proteins involved in the 3Rs are genome guardians that prevent a broad spectrum of diseases, ranging from cancers, immunological and neurodegenerative diseases, to developmental disorders such as diabetes and premature aging. Our lab investigates 3R mechanisms, 3R protein functions and structures, and their links to human disease. Our main research interests include:
- Mechanisms of genome duplication during growth and under stress conditions that mimics carcinogenic exposure or chemotherapeutic treatment.
- DNA repair processes that restore genetic information perturbed by a variety of DNA lesions arising from genotoxic exposures.
- The DNA damage response systems that monitor genome lesion burdens and deploy signaling pathways to induce multi-faceted physiological changes, ranging from metabolic and epigenetic changes, cell cycle delays, to chromosomal organization and maintenance adjustments.
To illuminate 3R processes and genome guardian functions, we have implemented cutting-edge and multi-disciplinary approaches, including genetic, biochemical, cell biological, structural, and genomic strategies. Our broad research scope and approaches provide lab members ample opportunities to explore and discover new biological principles. Our lab also offers a nurturing and supportive environment with tailored mentoring and training to advance lab members’ abilities in research, writing, presentation, and leadership.
Some of our research focuses driven by graduate students, postdocs, and undergraduate researchers in the 3R arena (Figure 1) are highlighted below. Please visit our publication site for further details.
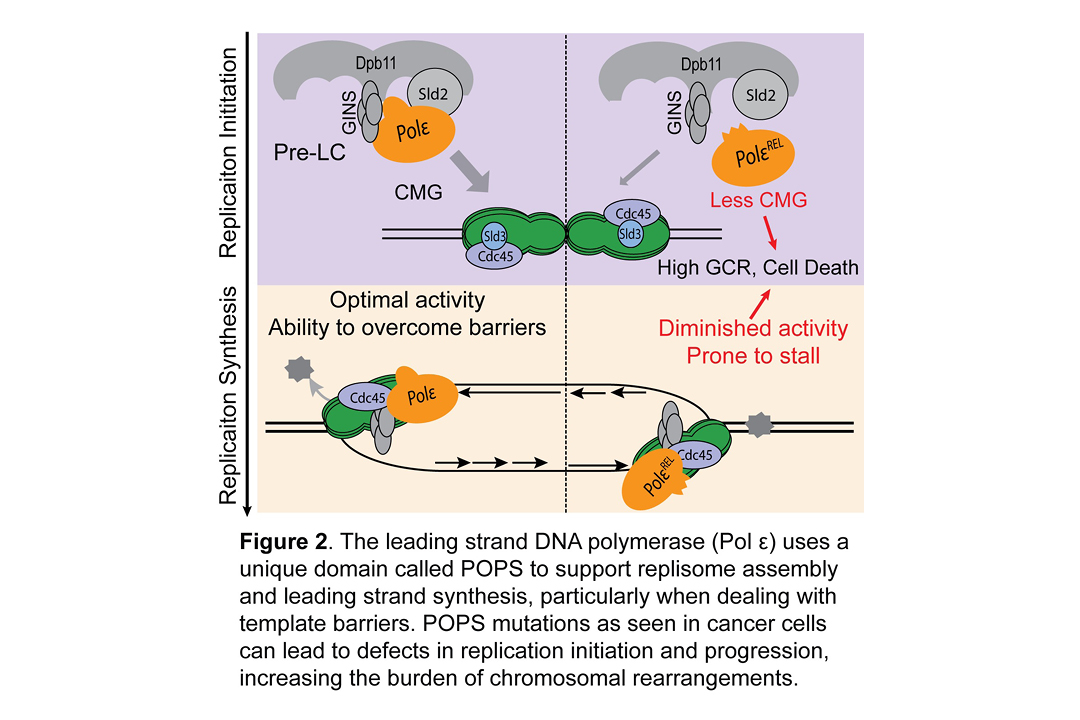
Mechanisms Governing Genome Duplication. Genome duplication is a complex and essential process that requires careful regulation during its three stages, namely replication initiation, progression, and termination. Our lab has made discoveries at each stage of replication. We revealed a novel SUMO-based control of the replicative helicase that prevents premature replication initiation in G1 phase (Wei and Zhao Nat Struct Mol Biol 2016). We identified a key feature that supports the leading strand polymerase in continuous synthesis of long stretches of DNA and linked this feature to cancer-prevention (Meng et al, Nat Commun 2020) (Figure 2). We uncovered a role for the essential Smc5/6 complex in completing replication (Peng et al PLoS Genet 2018). These studies were carried out by GSK and Tri-Institutional MD/PhD students and postdocs and they have opened up new avenues of further inquiries into the mechanisms governing replication and its regulation. For example, current lab members are uncovering additional SUMO-based control of replication initiation, new mechanisms to promote leading strand synthesis, and replisome dynamics from start to finish in the synthesis process.
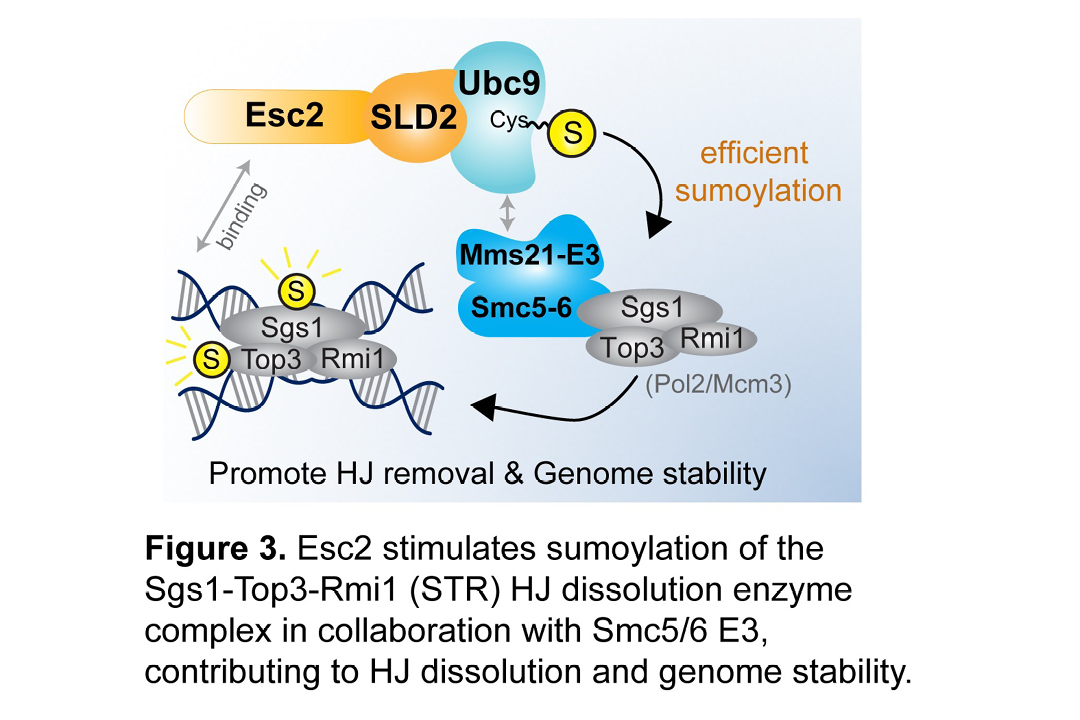
Recombinational Repair and Its Interface with Replication. DNA templates contain large numbers of barriers that impede replisome progression. Some examples of these barriers include DNA lesions generated endogenously or induced by carcinogens, DNA secondary structures, or transcriptional blockade. A large number of accessory proteins can help the replisome to overcome these barriers or fill ssDNA gaps left by failed replisomes. Recombinational repair proteins have roles in both processes. We found that some recombination enzymes can promote replication fork reversal, providing one way for replisomes to overcome barriers, and we showed that this activity is tightly regulated by the Smc5/6 complex (Xue et al Mol Cell 2014). Further, recombinational repair can fill ssDNA gaps and generate join DNA molecules called Holliday junctions (HJs). HJs must be processed into linear DNA for successful completion of repair. We revealed that HJ processing is controlled by multiple factors such as Smc5/6 and the Esc2 protein (Li et al Genes Dev 2021; Bonner et al Cell Rep 2016) (Figure 3) These discoveries made by BCMB PhD students and Postdocs have poised us to delve deeper into the interface between replication and recombinational repair.
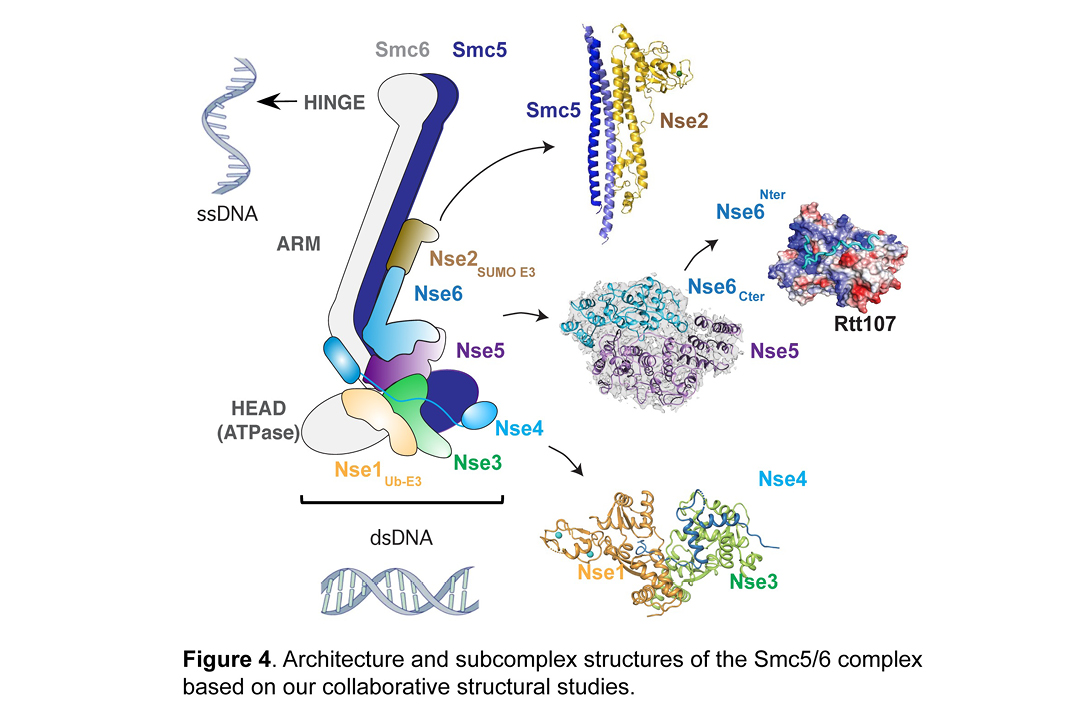
Smc5/6 Structures, Functions, and Disease Links. As the first group that identified the eight subunit Smc5/6 holo-complex, our lab has revealed that this complex ties together DNA replication and repair processes. Compared with the other two SMC (structural maintenance of chromosomes) complexes, namely cohesin and condensin that fold genomes, Smc5/6 has a more diverse range of capacities for modulating genome functions. We have provided biochemical insights into some of its activities. For example, we identified that Smc5/6 uniquely contains a SUMO ligase subunit, and provided structural explanation for this enzyme’s roles (Zhao and Blobel PNAS 2005, Duan et al Mol Cell 2009). More recently, we have integrated cryo-EM, chemical biology, structural modeling, and functional studies to reveal the architecture of the holo-Smc5/6 complex and atomic structures of its subcomplexes by assembling collaborative teams (Yu et al PNAS 2021, Jo et al JMB 2021) (Figure 4). We are continuing this endeavor to illustrate how this multiple-functional genome guardian contributes to DNA replication and repair and manipulate DNA. In addition, we are looking into mechanisms by which Smc5/6 acts as an important viral restriction factor that limit the replication and transcription of multiple cancer- and disease-causing viruses.
SUMO-Based DNA Damage Response. Modifying proteins via different modifiers is a principal means of regulating biological processes on a rapid time scale. SUMO modification can exert a dynamic range of effects on a protein’s functions, thus serving as an important switch or modulator of cellular processes. Focusing on the DNA damage response, we discovered that when cells are treated with agents mimicking chemotherapeutic drugs that induce DNA lesions, several dozens of 3R proteins become sumoylated. We termed this response “DNA damage-induced sumoylation” (DDIS) (Cremona et al Mol Cell 2012) (Figure 5). Since this discovery, we have been asking several mechanistic questions and provided insights into some of these questions. For example, we defined one mechanism by which cells sense DNA stress and initiate the DDIS (Chung and Zhao Gene Dev 2015). We established paradigms for how DDIS selects substrates and confers functional changes to enhance multiple DNA repair processes (reviewed in Sarangi and Zhao, Trends Biochem Sci 2015). We are currently researching on addition ways that DDIS is induced and achieve its effects in different molecular contexts.
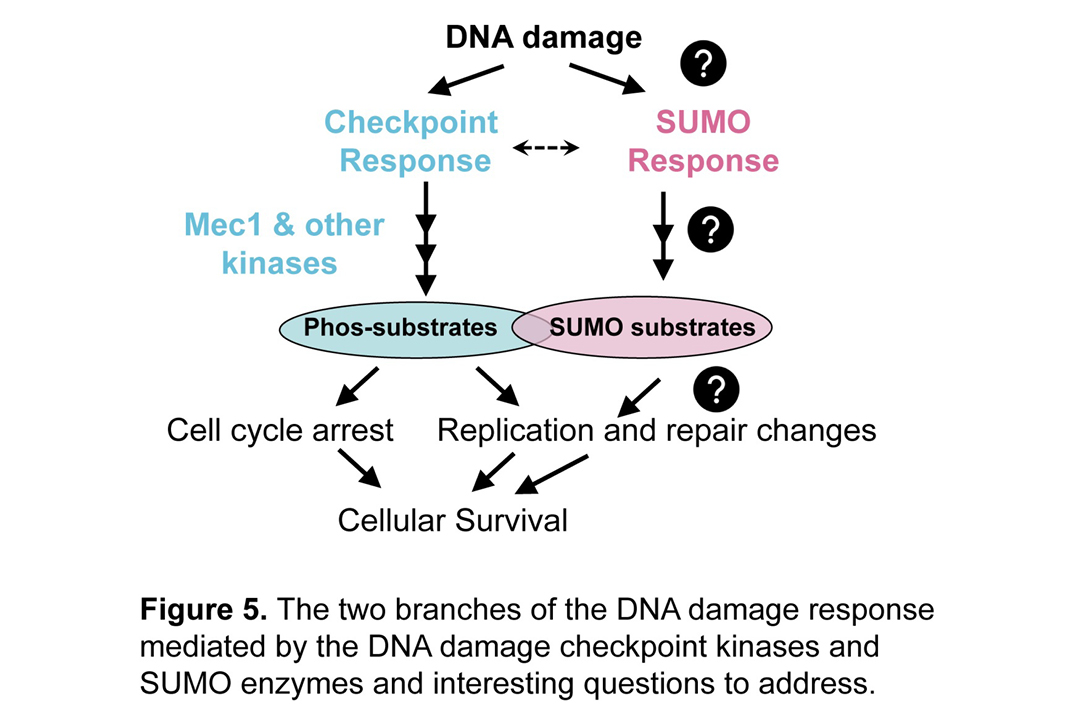
Kinase-based DNA Damage Response. We showed that DDIS operates largely independently the canonical kinase-based DNA damage response called the DNA damage checkpoint (DDC), suggesting the potential efficacy of a two-pronged approach to cancer therapy simultaneously targeting both DNA response branches (Figure 5). We are investigating the crosstalk between DDIS and DDC that can contribute greatly to a comprehensive view of the DNA damage response. In addition, we recently identified a new manner in which the cell can turn off the DDC after prolonged exposure to genotoxins, a process favoring proliferation and thus drug resistance (Dhingra et al PNAS 2021). Further research into these types of questions will provide a deeper understanding how DNA damage checkpoint operates and how cancer cells can short-circuit in favor of growth.
In summary, our lab integrates multidisciplinary approaches to study the genome maintenance processes that have important implications in tumorigenesis, viral infection, neurological diseases, and aging syndromes. Former students, postdocs, technicians, and undergraduate researchers from the lab have thrived in academics, medicine, biotech, and non-profit organizations. We welcome new members who are passionate about science and motivated to make impactful discoveries in a collaborative and fun environment.