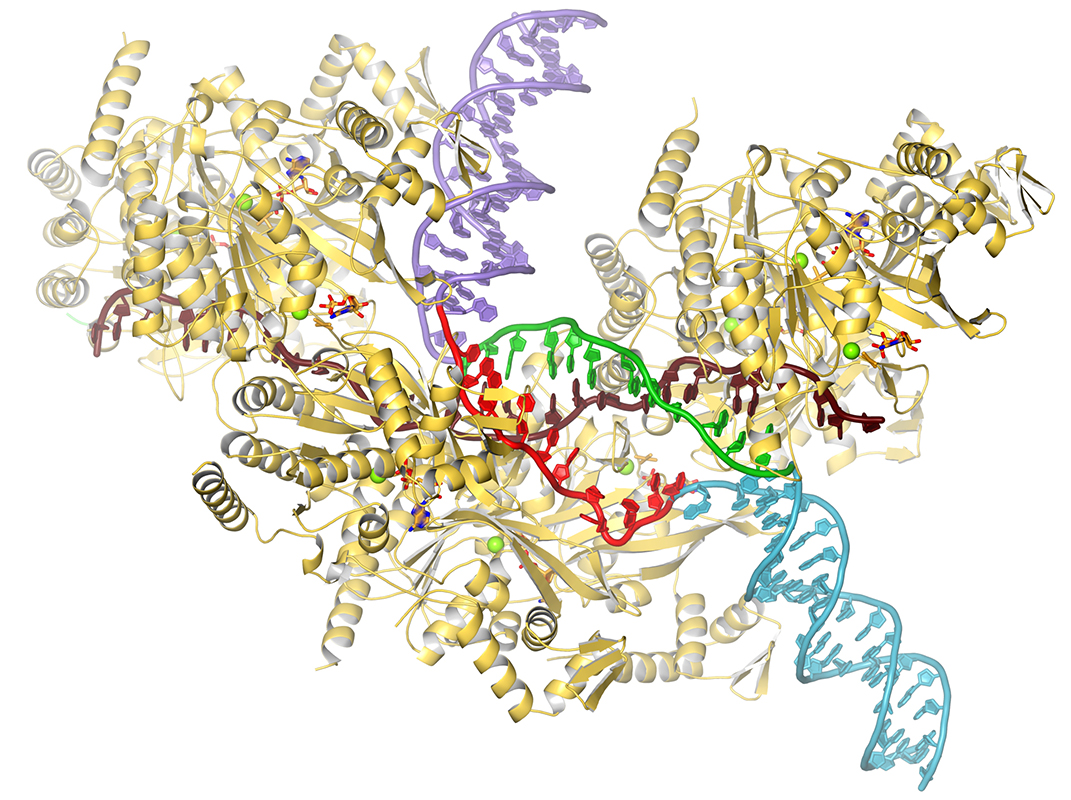
Homologous recombination plays essential roles in the repair of DNA double strand breaks (DSBs) and collapsed replication forks. The fundamental reaction in homologous recombination is the exchange of strands between a single-stranded DNA and a homologous double-stranded DNA. This reaction is carried out by the RecA/Rad51 family of ATPases. In repair, the DSB is resected to produce ssDNA, which is then coated by RecA/Rad51 to produce a presynaptic nucleoprotein filament, essentially the substrate-bound form of the recombinase enzyme (Chen et al., 2008). This filament then binds to dsDNA to form a synaptic filament, a key intermediate that mediates the search for homology and, when homology is detected, exchanges the DNA strands to produce a new dsDNA, termed heteroduplex. In a recent cryo-EM analysis of the recombination reaction (Yang et al., 2020) we found that the dsDNA strands are separated on binding to the filament. One strand is sequestrated while the other is freed to sample pairing with the ssDNA through Watson-Crick base pairing. Homology, through heteroduplex formation, promotes further dsDNA opening. Lack of homology suppresses it, keeping local synapses short. This presumably limits the futile strand separation in the absence of homology, allowing for the formation of multiple synapses to sample homology elsewhere along the dsDNA, increasing the probability of encountering homology. On ATP hydrolysis, which releases the DNA, a new heteroduplex is produced if strand exchange has occurred. Otherwise, the filament reforms for additional ATPase cycles of dsDNA binding, homology search and strand exchange until the correct register of the two DNA molecules is encountered.
This error-free form of DSB repair is crucial in preventing genomic abnormalities that can cause cancer. In fact, many of the proteins involved in the sensing, signaling and repair of DSBs were identified through studies of inherited cancer predisposition syndromes. These include the ATM and NBS1 proteins that are part of the DSB sensing complex, and the BRCA2 protein that binds to the resected ssDNA (Yang et al., 2002) and catalyzes the formation of the nucleoprotein filament by the RAD51 recombinase (Yang et al., 2005).