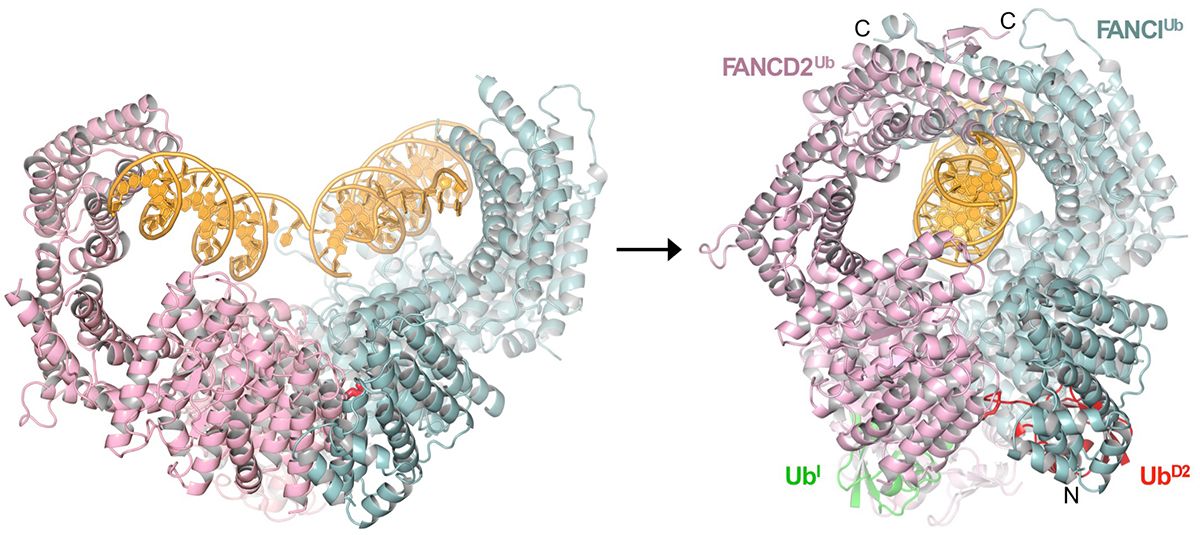
The Fanconi Anemia pathway, named after a cancer-predisposition syndrome, is essential for the repair of DNA-interstrand crosslinks (ICLs). The pathway consists of 22 FA complementation group proteins, which together with FA-associated proteins (FAAPs) and factors from other repair pathways are needed for the sensing, nucleolytic processing, translesion synthesis and homologous recombination-mediated repair of ICLs. A key event in pathway activation is the mono-ubiquitination of the complex consisting of the FANCI and FANCD2 paralogs (ID complex) by a nine-subunit ubiquitin ligase named FA Core complex. The crystal structure of the ID complex showed that it adopts an open trough-like structure with a basic interior that binds to DNA (Joo et al., 2011). However, the role of monoubiquitination was poorly understood. We addressed this question by determining the cryo-EM structure of the monoubiquitinated ID (IDUb) bound to DNA. IDUb adopts a closed ring structure that encircles the DNA. Compared to the cryo-EM structure of the non-ubiquitinated ID complex bound to ICL DNA, monoubiquitination triggers a complete re-arrangement of the open, trough-like ID structure through the ubiquitin of one protomer binding to the other protomer in a reciprocal fashion. In conjunction with biochemical data, the structure indicates that the monoubiquitinated ID complex looses its preference for branched DNA structures, becoming a sliding DNA clamp that can coordinate the subsequent repair reactions.
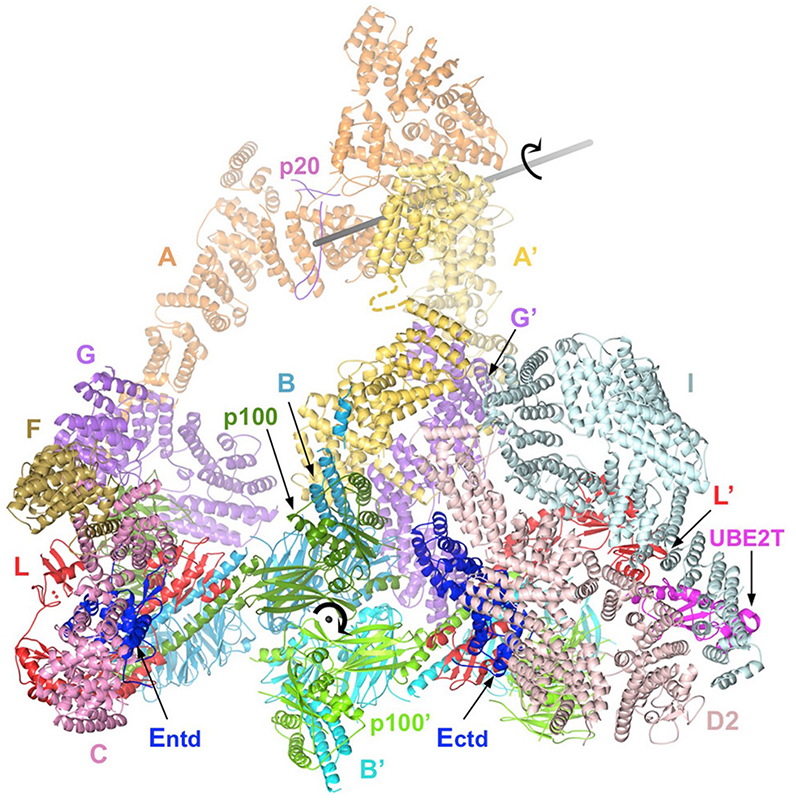
To understand how the ID complex is monoubiquitinated and how the conformational change to a closed clamp is triggered, we collected cryo-EM data on the Core-UBE2T-ID-DNA complex from a monoubiquitination reaction. We found that the complex adopts three distinct conformations during the reaction. The three structures, determined at up to 3.1 Å resolution, revealed that the Core-UBE2T complex remodels the ID-DNA complex to a closed-clamp conformation prior to ubiquitination, and the three conformations delineated the mechanism of clamp closing by the Core. We presume that subsequent monoubiquitination serves to prevent clamp opening after release from the Core. Together, these studies helped reveal the DNA clamp activity of the IDUb complex and the clamp-loader role of the Core complex.