A common way for science illustrators to depict genes is as segments of DNA located periodically along a stretch of chromosome, almost like station stops along a railroad.
But the reality is far more complex. Not only are the DNA sequences that make up a gene sometimes not continuous — not located at a single train stop — but genes also rely on regulatory regions of DNA, called enhancers, that can be many thousands of base-pairs away. In other words, if a gene starts at 30th Street Station in Philadelphia, its enhancer might be located in Boston’s Back Bay.
To transcribe a gene — convert it from DNA to messenger RNA on the way to making a protein — various cellular actors need to assemble at both the beginning of a gene (called the promoter region) while somehow also communicating with distant enhancers.
“A question that hasn’t really been resolved is how exactly this long-range gene regulation takes place,” says Alexandros Pertsinidis, a structural biologist at the Sloan Kettering Institute. “Somehow, the players have to come together to talk to each other, but how close they come, what the exact interactions are, and which molecules are involved is not really settled in the field.”
Many scientists believe that DNA’s flexible helix and protein regulators twist and fold together in some kind of local structure that permits transcription to take place. But that idea has mostly been speculative — until now.
In a new study published September 21 in the journal Nature Structural and Molecular Biology, Dr. Pertsinidis and his colleagues report for the first time detailed visual and biochemical evidence showing which features of DNA and proteins are necessary for a functional unit of transcriptional machinery to assemble in three-dimensional space in a cell.
Clusters, Droplets, and Networks
Clues to the blueprint of this nano-scale building project came about a year ago when Dr. Pertsinidis and a postdoctoral fellow in the lab, Jieru Li, observed tiny clusters of approximately 10 to 20 proteins congregating near the site of active gene transcription in a living cell. These clusters had never been observed before, likely in part because the necessary microscope power is hard to come by; Dr. Pertsinidis’s lab is a world leader in such nano-scale visualization.
But just seeing these clusters didn’t tell the scientists how they formed and what role, if any, they were playing in transcription.
To answer those questions, Dr. Pertsinidis and his team set out to systematically mutate — change the sequence of — various players involved in transcription to see whether these changes made a difference either to the assembly of the clusters or the occurrence of transcription. Another postdoctoral fellow in the lab, Lingling Cheng, joined on at this point in the project.
The scientists were hoping to be able to distinguish between two alternative models of how these clusters form.
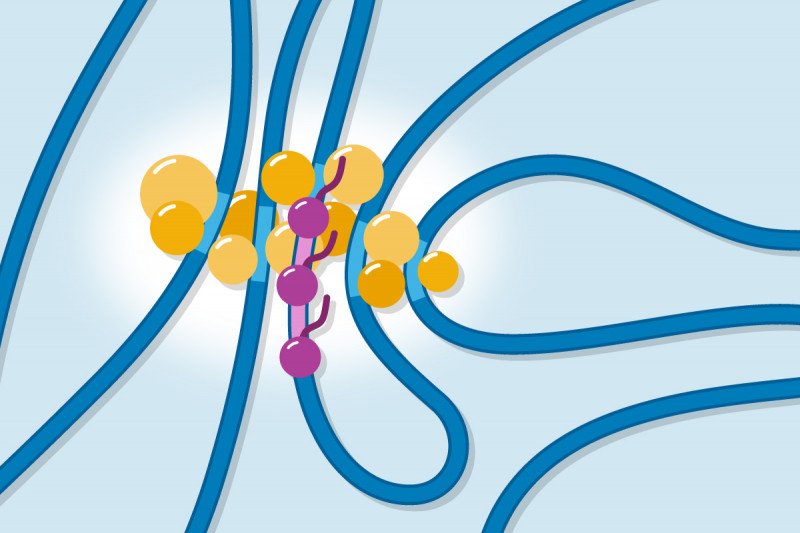
One model imagines that the regulatory proteins that perform transcription join up together through loose interactions to create “droplets” that surround and enfold the DNA. In this model, interactions between proteins are key to the formation of the clusters. And indeed, when scientists study these proteins in test tubes, they do form droplets under certain conditions.
The second model imagines that the regulatory proteins bind to specific regions of DNA (or chromatin, the proteins that wrap DNA) and it is this binding and the proximity of multiple such binding sites that facilitates the formation of the clusters.

The results of the experiments were clear and definitive: Altering the regions for the loose protein–protein interactions did not prevent the formation of the clusters. Changing the binding sites for protein–DNA and protein–chromatin interactions, however, did prevent cluster formation.
Therefore, the investigators concluded that proper communication between enhancers and target genes requires direct binding of regulatory proteins to these DNA and chromatin regions. By binding to multiple enhancers at one time, the regulatory proteins create a local network that brings the necessary players into close enough proximity to catalyze the reactions of transcription.
The results show the power of nano-scale microscopy. “Nobody had been able to zoom into a gene in a live cell to figure out where the various enhancers are located with respect to an active gene and also with respect to all the other regulatory factors that participate in transcription,” Dr. Pertsinidis says. “Now we have.”