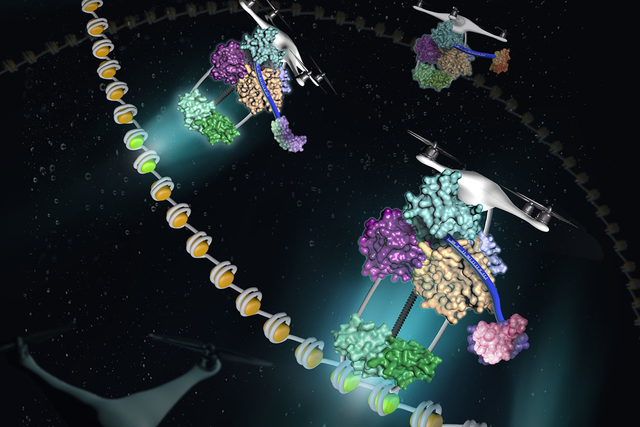
BRCT domains (colorful blobs) allow proteins to act like delivery drones, scanning metaphorical bar codes (blue strips) and carrying protein packages to the right spots along DNA molecules (gray coils).
One of the most significant scientific milestones of the 20th century was cell biologist Günter Blobel’s discovery of how proteins find their way to the right place in a cell. Tiny signal sequences within proteins act like postal codes for their destinations. One signal might specify mitochondria, while another might designate the nucleus.
These subcellular destinations, collectively called organelles, can be quite large. The nucleus, in particular, is practically a country of its own. Yet how proteins reach individual doorsteps within this vast territory is still mostly a mystery.

Molecular biologist Xiaolan Zhao
Researchers at the Sloan Kettering Institute have recently solved part of the puzzle. As they discuss in an article published on July 25 in the journal Molecular Cell, a secondary sorting system can carry proteins to distinct addresses inside the nucleus. A protein with repeats of a certain motif, called the BRCT domain, folds into the perfect shape so it performs like a drone of sorts, as if it is scanning a bar code and transporting cargo to the proper destination.
“This fold-and-scan transportation system can complement the general protein-sorting system to further route proteins to the right locations in the nucleus,” says Xiaolan Zhao, the molecular biologist at SKI who led the study. The findings contribute to fundamental knowledge in cell and molecular biology.
Two, Four, Six, Eight, BRCT Domains Congregate
Scientists first encountered BRCT domains in the cancer-predisposition gene BRCA1. (BRCT stands for BRCA1 C-terminal.) Mutations in the BRCT domain can hobble BRCA1’s function, which is to repair DNA damage. These mutations in the BRCT domain of BRCA1 prevent effective DNA repair and cause inherited cancer predisposition syndromes that are associated with increased risks of breast, ovarian, and other cancers.
Researchers now know that many proteins have BRCT domains. What unites these proteins is their participation in the crucial cellular process of maintaining genome integrity. DNA is constantly being damaged in different ways. A vast network of genome maintenance proteins is employed to repair DNA damage or find ways to copy DNA despite damage.
Many proteins, including BRCA1, contain two copies of the BRCT domain nestled side by side in the protein. This BRCT pair is dubbed di-BRCT. Its function is to bind phosphorylated versions of other proteins that become abundant when cells are exposed to UV light, carcinogens, and DNA-damaging cancer drugs. (Phosphorylation is the process of adding phosphates to molecules.) “These phosphorylation events are a sign of DNA damage,” Dr. Zhao says. “They’re telling the DNA maintenance network, ‘We are under stress. You should do something.’ ”
Proteins can have more than two BRCT domains. The SKI researchers were interested in the protein RTT107, which has six: two at one end and four at the other. What are the four BRCT domains doing, they wondered? Do they act in the same manner as the di-BRCT found in BRCA1, only doubled? Or do they take on some new function?
Into the Groove
A postdoctoral research fellow in Dr. Zhao’s lab, Bingbing Wan, set out to solve the structure of the four-BRCT region. He discovered that it has a completely different shape than two independently folding di-BRCT domains. The four BRCT domains fold together into a tetrahedral (four-sided) shape with a deep groove running through one side. This feature suggests that proteins might attach themselves to RTT107 by snuggling inside this groove. The researchers call this unique type of folding “molecular origami.”
RTT107 partners with several proteins that are vital to life. These proteins, it turns out, all share a similar amino acid sequence that allows them to fit inside RTT107’s groove. The interaction between RTT107 and each of theses proteins in this consensus sequence is very specific and mutually exclusive.
Dr. Wan likens the consensus sequence to a bar code that RTT107 can read. “The bar code tells the delivery vehicle where each package is going,” he says.
A Molecular Tether
Next, the investigators asked how the four-BRCT section of RTT107 cooperates with the di-BRCT section. Di-BRCT domains were already known to bind to phosphorylated histones. (Histones are proteins around which DNA is wrapped like yarn on a spool.) The team hypothesized that RTT107 serves as a molecular tether between these histones and the proteins to which the four-BRCT domain binds. In this model, di-BRCT binds to phosphorylated histones, while the four-BRCT binds to genome maintenance proteins, helping them find their way to a specific region of DNA.
The team’s data support their hypothesis. They further found that mutations interrupting the association between genome maintenance proteins and RTT107 impair DNA duplication and generate large-scale DNA rearrangements. These types of changes can fuel cancer growth.
Many BRCT domain–containing proteins are what are called scaffolding proteins. They don’t have enzymatic activity themselves but rather act as organizational hubs. They bring together multiple proteins to execute a common task. RTT107 is one example.
Dr. Zhao says that these findings may apply to other scaffolding proteins. “It will be interesting to find out if other proteins can act as secondary sorters for delivering genome maintenance factors to different parts of DNA,” she says.