No technology has had a greater impact on biology in recent years than the genome-editing tool CRISPR. With these molecular scissors, researchers can make precise changes to DNA at very specific parts of a genome — say, insert a new gene, edit a gene’s sequence, or even remove a section of DNA altogether.
Compared with previous tools, CRISPR is also faster, easier, and cheaper.
But the CRISPR tool is only as good as your knowledge of the genome you are cutting. If the genome has multiple targets for CRISPR’s sheers, then instead of making precise snips in the genome, you might make Swiss cheese of it.
This is the conclusion of a new study by a team of researchers in the Sloan Kettering Institute (SKI), who also present a way to solve the problem. They have developed a software program that raises the bar on CRISPR’s precision, making it much more reliable as a genome-editing tool.
A Bacterial Cut-and-Paste System
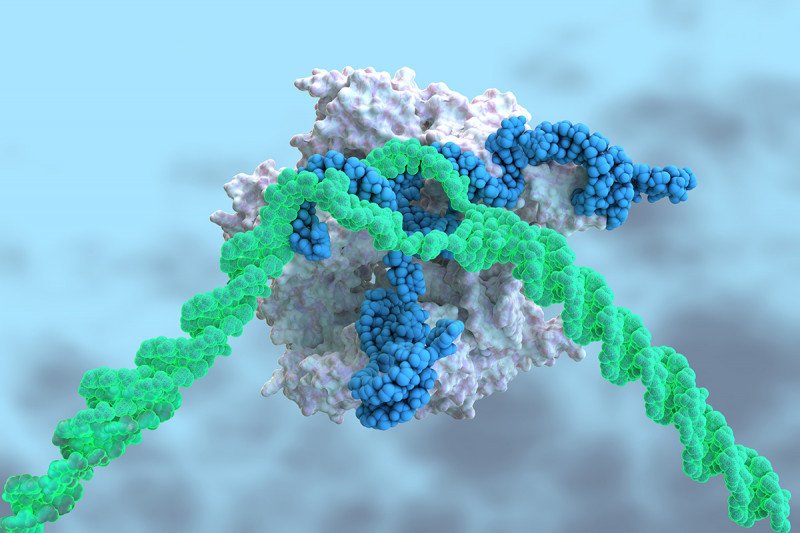
CRISPR stands for clustered regularly interspaced short palindromic repeats. These short sequences of repetitive DNA are found in bacteria and other microorganisms. Microbes use these sequences as a type of immune system against viruses.
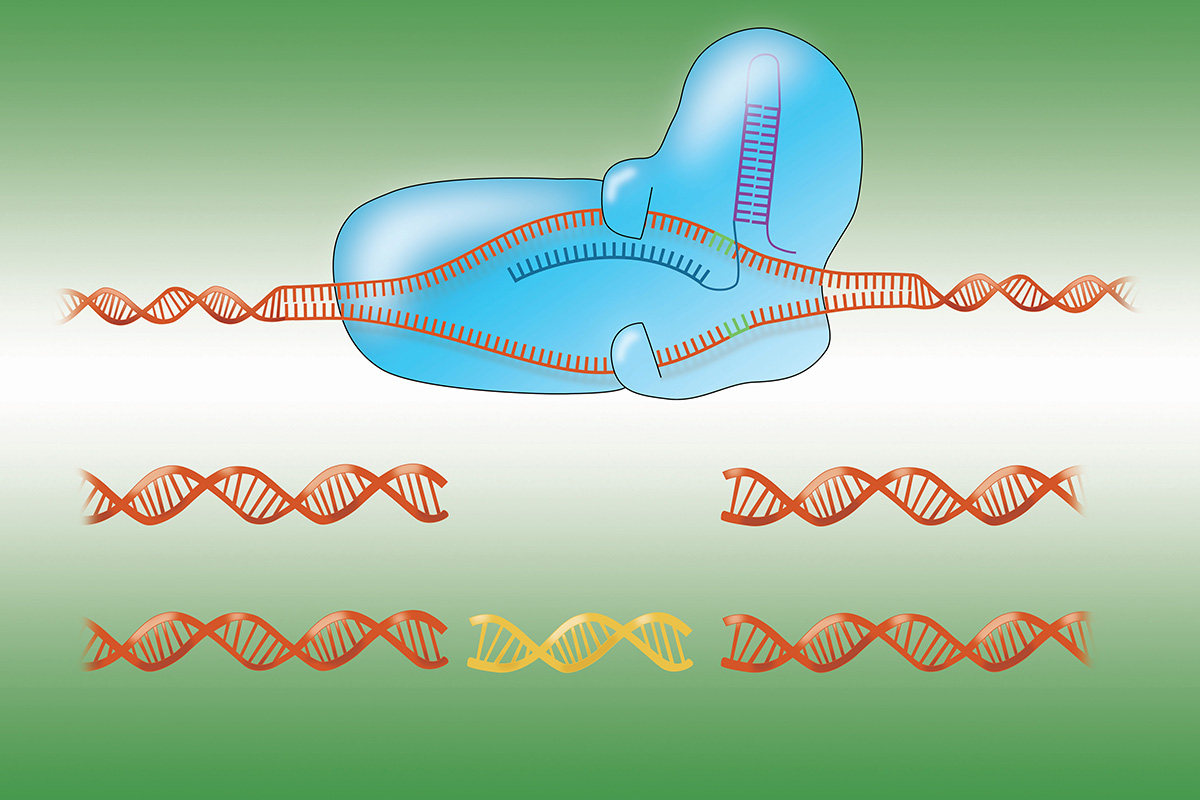
Key to the whole operation are stretches of DNA in the CRISPR sequences that match the genetic sequence of viral DNA. When these sequences are transcribed into RNA, they bind to viral DNA and direct an enzyme — for example, one called Cas9 — to snip the DNA at the match site. This disarms the invader. Bacteria retain a record of past viral infections, much like our own immune system does, stitched into their DNA.
In 2012, scientists at the University of California, Berkeley, and elsewhere realized they could turn this bacterial cut-and-paste system into a powerful tool for genetic engineering. Researchers can give the Cas9 enzyme a synthetic RNA as a guide, one that matches a gene or DNA sequence of interest. They can then use the CRISPR-Cas9 system to make very precise cuts and other alterations in the DNA of cells.
Know Thy Genome
Now comes the tricky part. When researchers design their guide RNA, they often do so without full knowledge of where in the genome that guide might bind besides the specific region of interest. Genomes are big, and just by chance it’s possible for the guide RNA to match more than one genome location. That’s bad if what you are trying to do is limit a genetic change to one specific region. Without a way to eliminate off-target matches, you’re liable to make unwanted genetic changes.
“You always have to have a picture of the whole genome if you want to know how specific your guide RNA is,” says Joana Vidigal, a postdoctoral fellow in SKI. She is a co-author on a new paper in Nature Biotechnology that addresses this problem.
She and her colleagues, including SKI postdoctoral fellow Yuri Pritykin and graduate student Alexendar Perez, built a software program that, with a click of button, gives researchers a list of all the unique guide RNAs for a given genome. When researchers use this software, called GuideScan, they can be sure that their guide RNA will not bind anywhere else in the genome.
That’s not the case with other available tools, explains Christina Leslie, a computational biologist in SKI and a senior author on the paper. “There could be hundreds of perfect matches in the genome with these other tools,” she notes.
Some tools will even give you a score that is supposed to reflect how specific the guide RNA is. But since they don’t take into account all potential off-target sites, Dr. Leslie points out, “they’re not meaningful scores.”
“You could be shredding your genome because you think that you’re editing in just one place but there are actually all sorts of cuts,” Dr. Vidigal adds.
With GuideScan, on the other hand, researchers know that every guide RNA in the database will have only one perfect hit. “We can provide some guarantees,” Dr. Leslie says.
Greater Coverage
Another plus of the new approach is that the software covers noncoding regions of the genome. Noncoding regions are those that don’t contain information for making proteins. Researchers used to think of them as junk DNA, but they now know these regions are anything but. Noncoding regions contain sequences that control when, where, and how much of a protein is made. Sometimes researchers want to make changes in these regions, too — say, cut out a regulatory sequence and see what happens to the cell as a result.
Such expanded genome coverage permits the easy creation of a type of mutation that is especially common in cancer, called a translocation. This is when big chunks of DNA move from one place to another, often fusing two genes together as a result. Before CRISPR, it was extremely tedious and difficult to make these mutations. Now, all you need is two guide RNAs that match the desired breaking points. The cell’s DNA repair system will join these two otherwise distant pieces of chromosome.
“CRISPR has completely changed the way we do this type of research,” says Andrea Ventura, a cancer biologist in SKI and a co-lead author on the study. “You can actually create these mutations at a fraction of the cost and in a much more straightforward and elegant way.”
Several years ago, Dr. Ventura’s lab used the technology to create this type of mutation in lung cells in mice and reliably induce cancer, thus creating a useful model for study.
But that is really just the beginning, says Dr. Vidigal. “Pretty much anything you can imagine you can do. And now you can do it precisely.”