We are interested in the mechanism of chromosome replication, a process that is highly conserved across eukaryotes and that involves the duplication of both the chromosomal DNA and its associated chromatin states. As chromosomes are the carriers of both the genetic and epigenetic information, faithful chromosome replication is of fundamental importance for genome maintenance during normal cell proliferation. Accordingly, defects in chromosome replication are a major driver of the genomic instability observed in cancer cells. To understand the molecular mechanisms by which eukaryotic cells carry out and monitor accurate genome replication we employ a fully reconstituted DNA replication system based on purified proteins from the budding yeast, S. cerevisiae. Research projects are focused on the mechanistic characterization of the DNA replication machinery, the mechanism of replication-coupled chromatin assembly, and the control of DNA replication by checkpoint genome surveillance pathways.
Research Projects
DNA replication and replication stress
In eukaryotes, DNA replication initiates at many replication origins that are distributed along the length of each chromosome arm. At each origin, two replication forks are established that travel in opposite directions to form a replication bubble (Figure 1).
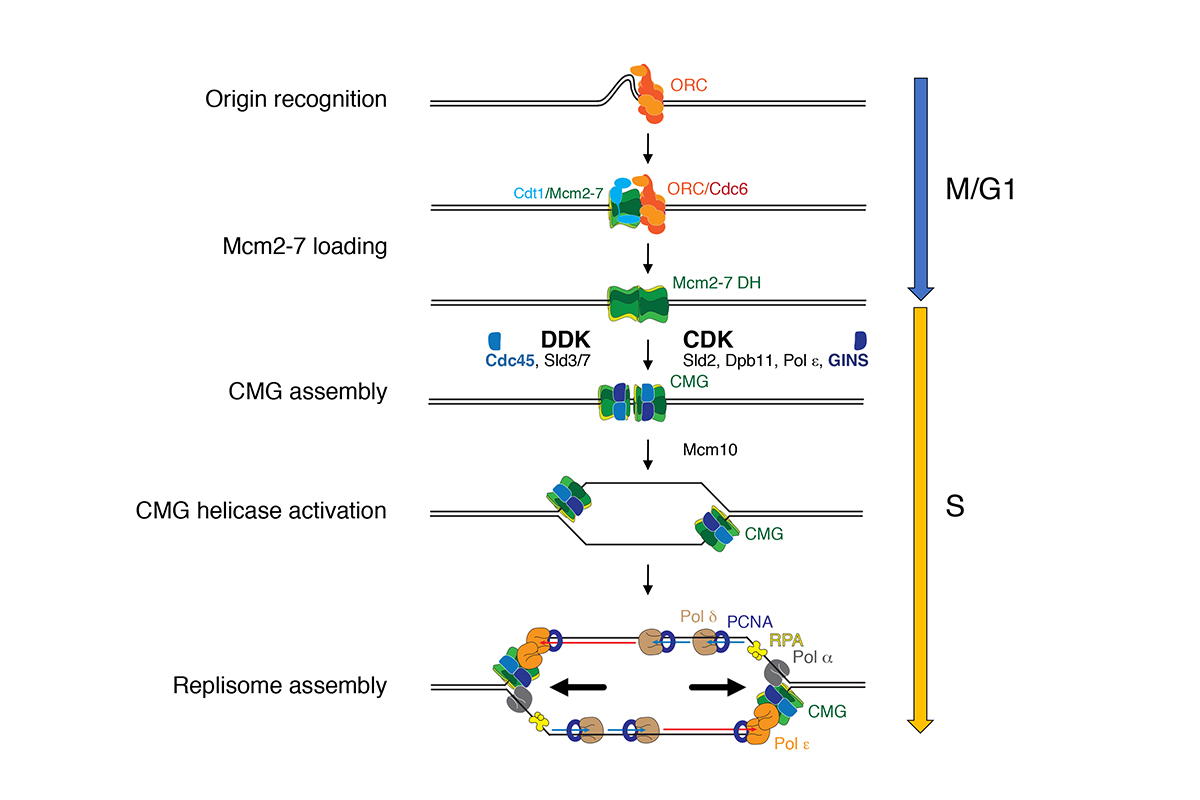
Figure 1: Mechanism of DNA replication in eukaryotes.
A large multi-subunit protein complex, called replisome, coordinates DNA unwinding with DNA synthesis at the fork. Over 20 distinct proteins and protein complexes, comprising over 50 gene products, are minimally required to establish replisomes at eukaryotic replication origins. We have reconstituted this process with purified proteins from the budding yeast, Saccharomyces cerevisiae, which has allowed us to define mechanisms involved in origin site selection, origin activation, and fork progression (Figure 2).
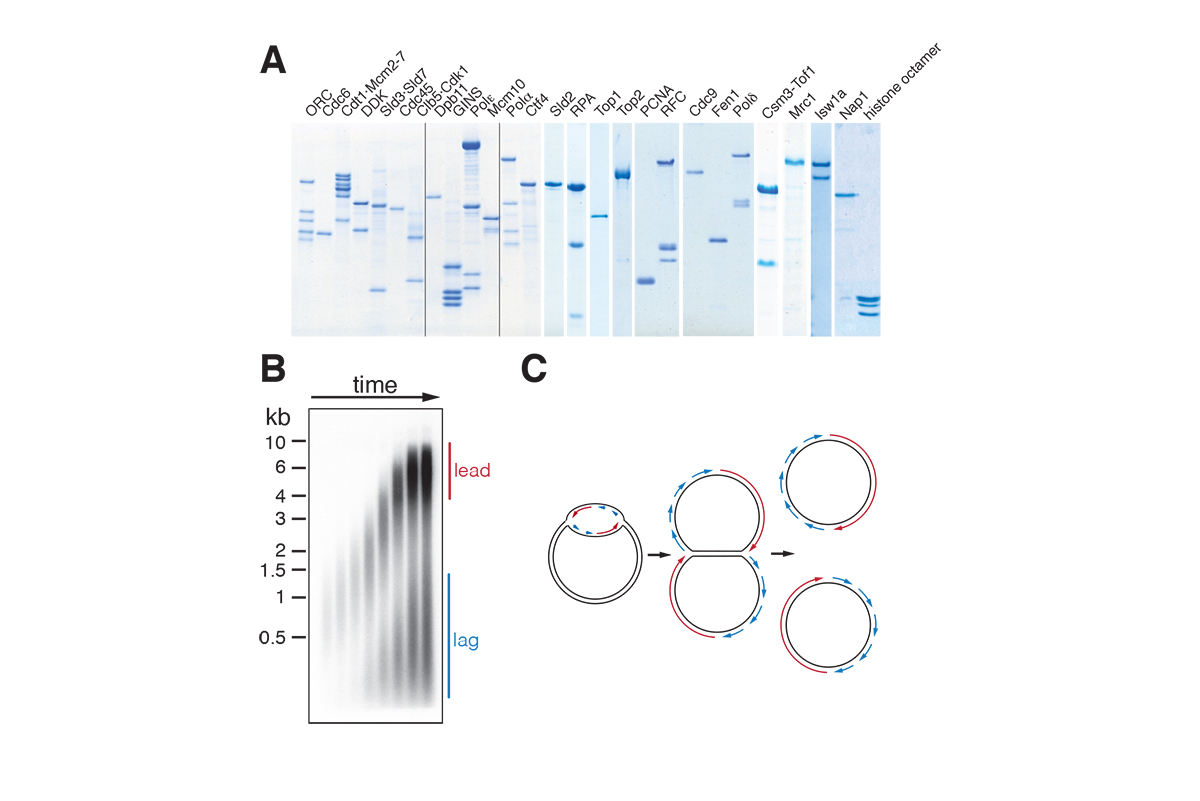
Figure 2: Reconstitution of DNA replication with purified proteins (A) Purified replication proteins. (B) Denaturing gel analysis of plasmid replication products. (C) Plasmid replication model.
Using this system, we are interested in addressing fundamental questions on DNA replication. For example: How are two replication forks of opposite polarity established at replication origins? What is the molecular function of the many accessory proteins that promote fork assembly and progression? Moreover, we are interested in characterizing the molecular nature of replication stress: How does dNTP depletion, a prominent consequence of oncogene-induced replication stress, impact DNA replication fork progression? How do physical obstacles on the DNA template, such as non-nucleosomal protein-DNA complexes or R-loops, affect fork progression? How do accessory genome maintenance factors, such as DNA helicases, promote fork passage past physical obstacles?
DNA damage and DNA replication checkpoints
The S phase checkpoint is a kinase signaling cascade that is activated in response to DNA damage or fork stalling during replication stress. Checkpoint activation results in the stabilization of stalled forks, prevents further origin firing, and creates a cellular milieu that promotes the resolution of replication fork blocks before entry into mitosis by inducing cell cycle arrest, dNTP production, and the transcription of DNA repair factors. In previous work, using the reconstituted DNA replication system, we have identified the molecular functions for the effector kinase, Rad53, in controlling fork progression and origin activation by inhibiting the activities of the replicative DNA helicase, CMG (Cdc45-MCM-GINS), and the cell cycle-regulated protein kinase, DDK (Dbf4-dependent Cdc7 kinase), respectively (Figure 3).
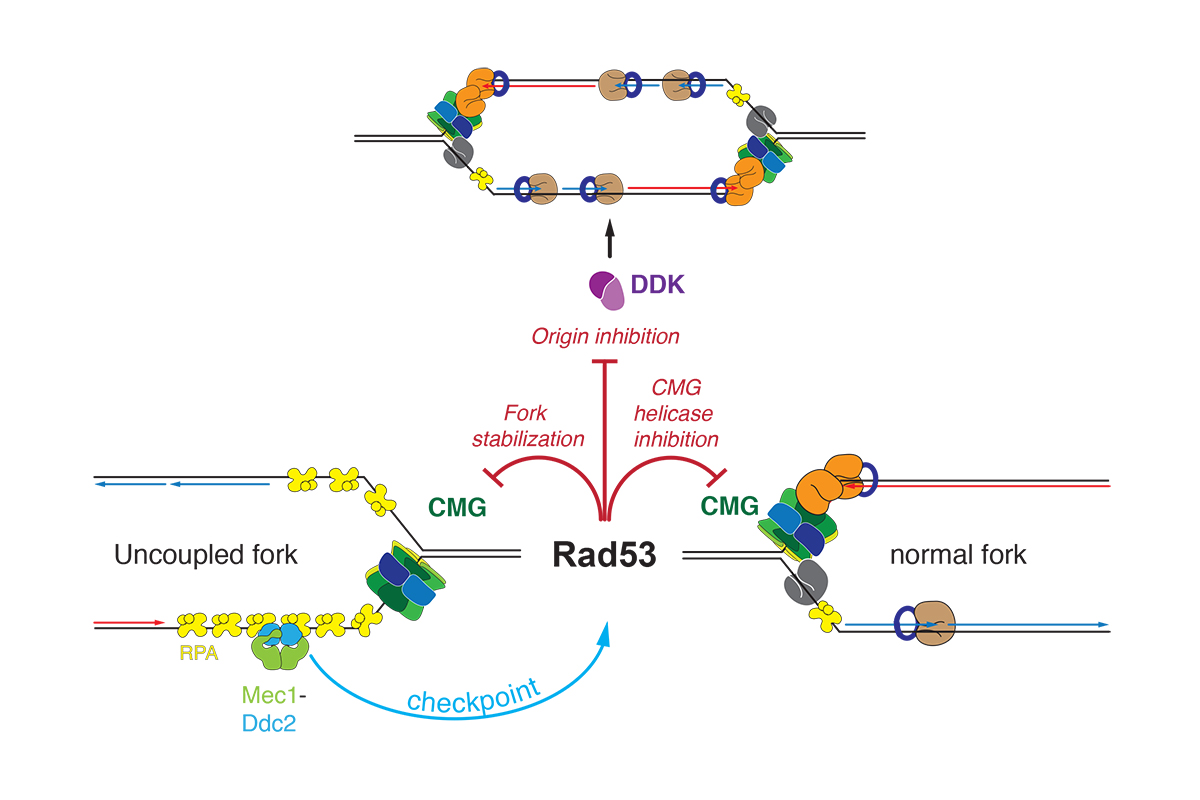
Figure 3: Checkpoint regulation of DNA replication.
In ongoing work, we aim to elucidate the molecular mechanisms of these Rad53 functions. Rad53 activation is controlled by the activation of the apical checkpoint kinases Mec1 / Tel1 (ATR / ATM in humans) and various mediator proteins that transmit the activation signal from the sites of stalled forks or DNA damage. We are interested in elucidating the mechanism of the checkpoint signaling cascade using biochemical and structural approaches.
Replication-coupled chromatin assembly
Nucleosome positions and histone modifications are critical determinants of a cell’s gene expression program and, therefore, cell identity. However, the process of DNA replication intrinsically disrupts nucleosomes ahead of the fork raising the question how chromatin structure is maintained across generations. Nucleosomes are re-assembled on the daughter chromosomes behind the replication fork from both parental nucleosomes and de novo from newly synthesized histones. Intriguingly, several replisome components, such as subunits of the DNA helicase and DNA polymerases, exhibit histone chaperone activity that is important for the normal segregation of parental histones at the fork. In addition, a host of additional histone chaperone complexes and chromatin remodelers is known to be required for the assembly and maturation of nascent chromatin (Figure 4).
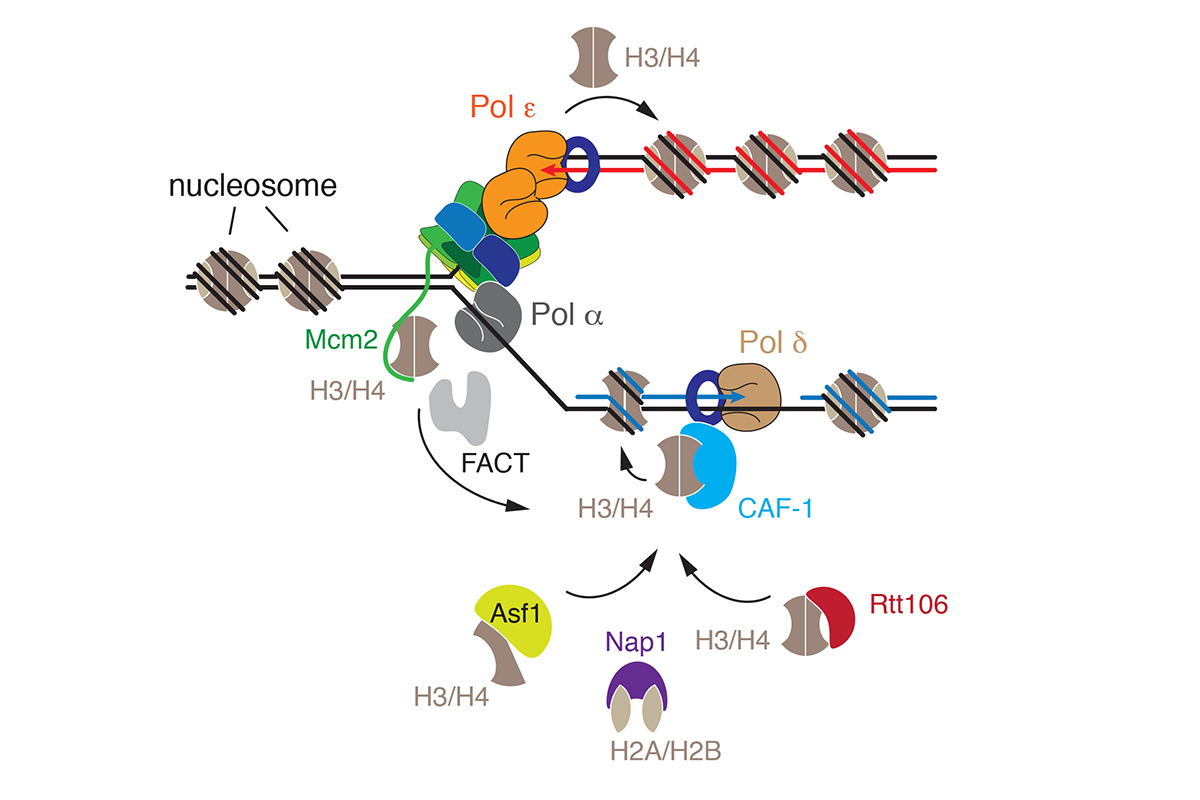
Figure 4: Replication-coupled chromatin assembly.
We are interested in determining how replication forks navigate the chromatin landscape, how parental nucleosomes are physically recycled from the front of the fork to the daughter DNA strands behind the fork, and how replication-coupled chromatin assembly factors integrate with the replication machinery to re-assemble chromatin on newly synthesized DNA.