Homologous recombination (HR) and nonhomologous end- joining (NHEJ) are considered the two primary DNA double strand break repair (DSBR) pathways. However, there are a number of secondary DSBR pathways, used at low levels in all cells, but are particularly relied upon in cancer cells.
Alternative end-joining makes use of available short stretches of complimentary base pairs on the 3’ single stranded overhang formed through end-resection. Also known as microhomology-mediated end-joining, Alt-EJ is incompletely understood since only 50% of events are mediated through the only known Alt-EJ specific factor polymerase theta.
The focus of our lab is to contextualize the use of Alt-EJ relative to other pathways in cancer and normal cells by understanding genetic and other influences that promote or diminish its use.
HPV oncoproteins and DNA repair pathway deficiencies
HPV-associated squamous cell carcinomas are highly radiosensitive when compared to their HPV negative comparators, a phenomenon that is consistent for aerodigestive and anogenital cancers. Of all biomarkers in radiation oncology, HPV positivity is uniquely well validated and possibly clinically actionable through radiation dose de-escalation. Our group characterized the DNA double strand break defect in HPV-associated cancers using three methodologies and found corroborating evidence by identifying a microhomology-based indel genomic signature in HPV-associated cancer genomes reflective of increased Alt-EJ usage. We continue to study basic mechanisms of how HPV oncoproteins affect DNA repair capacity and pathway choice.
Technology development: improving measurement of primary and alterative DSB repair pathways
With support from the NCI and the Innovative Molecular Analysis Technology (IMAT) program, we develop innovative new tools to measure both primary and secondary DNA DSB repair pathways by comprehensively profiling insertions and deletions of all sizes. With support from the Emerson Collective Cancer Research Fund, we apply these tools to human specimens to measure repair capacity in human tumors.
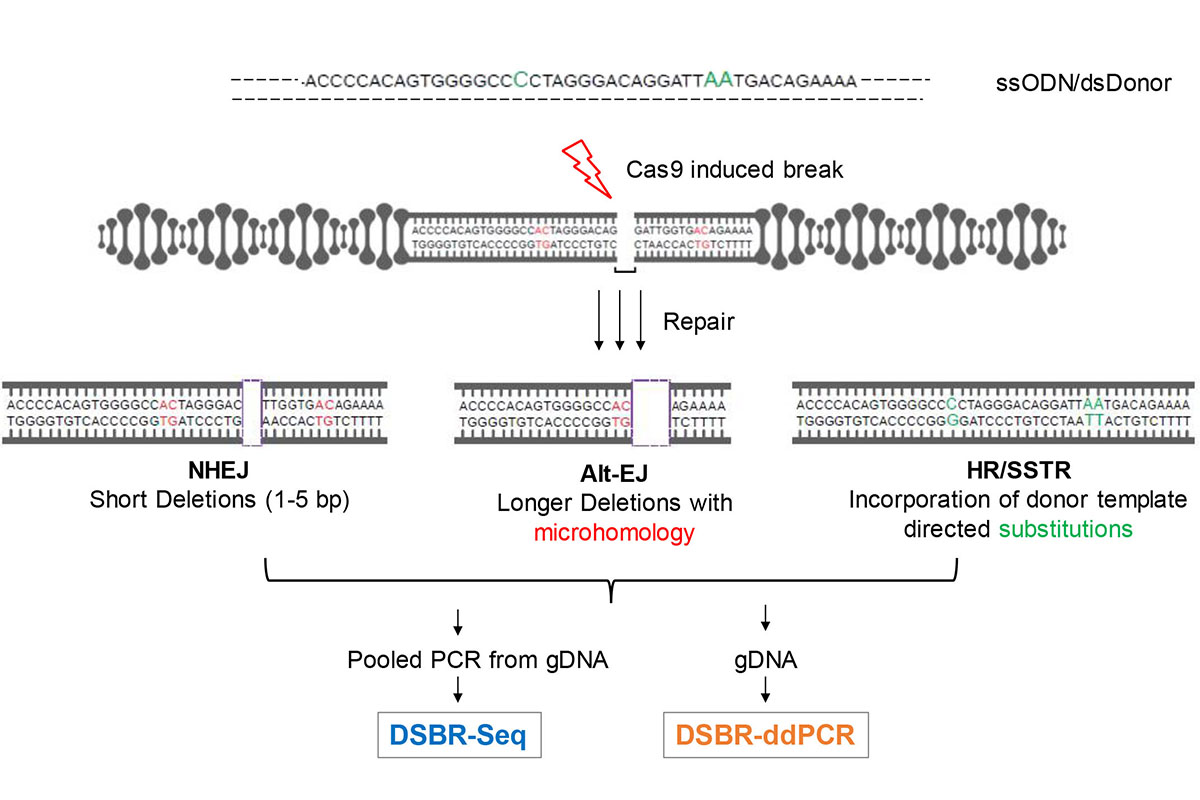
Figure 1. Schema of approach. Cas9 and gRNA expression plasmids are transfected into a cell population with or without a DNA donor sequence. Both single stranded oligonucleotide donors (ssODN) and double stranded DNA donors (dsDonor) with 3 substitutions (in green) are used in this study. Following an incubation period of typically 2-3 days, a range of repair pathway usage are assessed using PCR amplication of a 201 bp segment around the break followed by sequencing of the PCR product with paired end 150 x 2 Illumina MiSeq sequencing (DSBR-seq). Alternatively, genomic DNA is used as an input for droplet digit PCR(ddPCR) to measure specific repair products on a Biorad QX200 platform (DSBR-ddPCR).
End-resection, end protection and Alt-EJ regulation
Alt-EJ is at least partly mediated through a factor known as polymerase theta, which mediates annealing and repair of short segments of microhomology typically located within 15 bp from the breaksite. End-resection is highly regulated with multiple mechanisms to limit resection to S and G2 portions of the cell cycle when complementary sister chromatid DNA is available for homologous recombination. However, Alt-EJ requires only short-range resection in contrast to HR. In addition, the 53BP1-Shieldin axis protects ends from resection, but apparently requires exposed single strand DNA to operate. Using optogenetic control of Cas9-induced breaks, we study how short range resection, end protection, and cell cycle phase regulate Alt-EJ utilization.
Matching radiosensitizing DNA damage response inhibitors(DDRi) to repair deficiencies in cancer
An array of small molecular inhibitors targeting components of the DNA damage response have entered phase I clinical trials, including inhibitors of the ATM, ATR, and DNApkcs kinases. Though clearly radiosensitizing, it is not clear how these agents might improve the therapeutic window of responses to radiation between tumor and normal cells. Using repair profiling of DNA repair capacity, we seek to find novel categories of tumors with improved therapeutic windows when matched to an optimized DDRi.