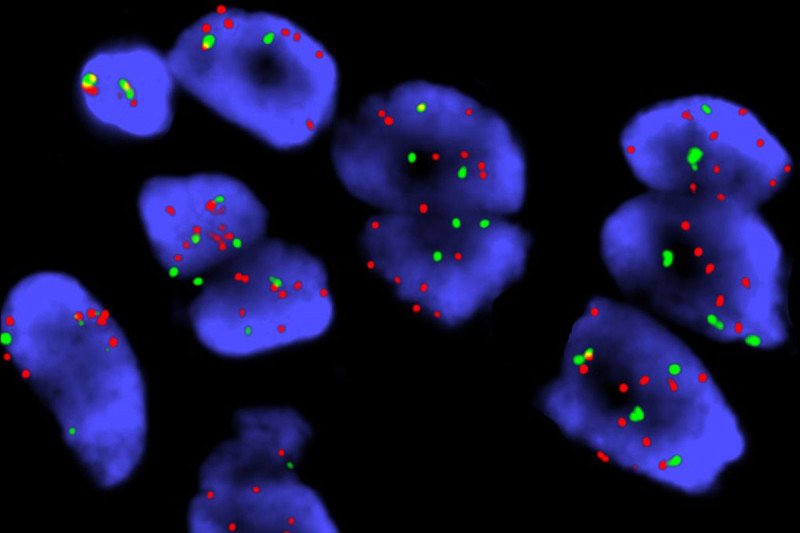
This image shows cells taken from a person with lung cancer and labeled with fluorescent probes (a technique called FISH). The red probe is specific for the YES1 gene. The green probe recognizes a part of chromosome 18 where YES1 is usually found. Cells with extra copies of the YES1 gene have significantly more red spots than green spots. Image courtesy Lu Wang.
In the era of precision medicine, people with identifiable mutations in cancer-related genes can be considered both lucky and unlucky. Knowing which genes are driving a cancer gives doctors a clear target for therapies, and patients generally benefit immediately.
The downside is that these drugs tend to work only temporarily, as cancers eventually evolve ways to get around them.
A type of lung cancer that tends to affect nonsmokers, for example, is exquisitely sensitive to drugs that block a protein called EGFR; even very large tumors can rapidly melt away after treatment.
Unfortunately, nearly all people treated with these drugs will eventually develop resistance to them. When that happens, the tumors come back.
“The problem of therapeutic resistance is universal across oncology,” says Pang Fan, a physician-scientist in the Human Oncology and Pathogenesis Program at MSK. “Our goal is to try to identify mechanisms that underlie the emergence of resistance, so that hopefully we can develop ways to overcome them.”
Dr. Fan is the first author on a paper that identifies a new mechanism of resistance to EGFR inhibitors in people with lung cancer. The innovative study — a collaboration between scientists at MSK and the New York Genome Center (NYGC) — suggests treatment options for people who develop this form of resistant disease.
Making Mutations with Mobile DNA
To identify genes involved in resistance, the team used something called transposon mutagenesis. Transposons are bits of DNA whose specialty is being able to pick up and move from one region of the genome to another. These mobile elements can be deliberately introduced into cells to produce mutations in different parts of the genome — wherever they land, essentially.
The scientists introduced the transposons into a cell line that is sensitive to EGFR inhibitors. Then, they grew the cells in dishes along with an EGFR-inhibiting drug. The cells that were able to survive and grow in the presence of the drug — presumably those with resistance mutations — were then harvested, and their DNA was extracted.
By comparing which mutations occurred repeatedly across different experiments, the team was able to home in on one gene that seemed to confer resistance when it was turned on. That gene is YES1.
This gene makes a protein called a kinase, one belonging to the src family of kinases.
Once they knew that YES1 was involved, the team then looked to see if this mutation is also common in people who have received EGFR inhibitors. They used MSK’s cBio Portal, which houses the results of MSK’s genomic sequencing test, MSK-IMPACTTM. They identified 136 samples obtained from people with lung cancer after their disease was no longer kept in check by EGFR drugs. A description of these samples was recently published in the journal Clinical Cancer Research. Notably, four of them had gained extra copies of the YES1 gene.
“It’s not a large fraction of patients who develop this resistance mechanism,” Dr. Fan says. “But keep in mind that mutations in EGFR drive the growth of about 20 to 30% of all lung adenocarcinomas.”
Even better, drugs that target this family of kinases are readily available and already approved by the US Food and Drug Administration for other cancers, although clinical trials would be needed to establish their efficacy for this new indication.
A Productive Collaboration
Dr. Fan started this research as a postdoctoral fellow in the lab of Harold Varmus at MSK, before Dr. Varmus left to become head of the National Cancer Institute. (Dr. Varmus is now the Lewis Thomas University Professor of Medicine at Weill Cornell Medicine and a Senior Associate Core Member at the NYGC.) Dr. Fan then completed the work in Marc Ladanyi’s lab at MSK. Dr. Ladanyi is Chief of the Molecular Diagnostics Service.
The NYGC was critical for making sense of the reams of transposon-altered genomes, Dr. Fan explains. “They figured out what would be the most effective way to process these experimental samples, then performed the next-generation sequencing and customized the software to analyze the sequencing data,” he says.
“This study is a good example of how combining powerful laboratory tools and our institutional clinical genomics effort, of which MSK-IMPACT is the centerpiece, can generate compelling data that could affect patient care,” Dr. Ladanyi adds. “We plan to continue this collaboration with the New York Genome Center to better define mechanisms of resistance to other targeted kinase inhibitors used in patients with lung cancer.”
The study was published in the journal Proceedings of the National Academy of Sciences.







