Single cells from the embryo of a mouse are plotted according to their similarity in gene expression. Each dot represents the expression of a particular gene in one cell. Different colors correspond to different types of cells in the embryo. Single-cell analysis like this is a powerful tool that allows researchers in the Sloan Kettering Institute to paint a clearer picture of development, one cell at a time. Image: Courtesy of the Hadjantonakis lab
Tissues in the human body are like a pointillist painting. From afar, they appear smooth and seamless, but up close, millions of individual cells reveal distinct identities.
A cell’s identity is determined by the genes it turns on. Until recently, the only way to learn what genes were turned on in cells was to look at the blend of genetic material pulled from thousands of cells. This technique provides a clear measure of overall gene activity but is a poor indicator of what’s happening inside individual cells.
And that’s a problem because in biology, even one cell can make a difference.
Thanks to an approach called single-cell analysis, the picture is changing. This technique combines advanced genomic sequencing tools and sophisticated computational methods. With it, biologists can peer inside individual cells to see which particular genes are turned on.

Developmental biologist Kat Hadjantonakis (left) and her team are helping to answer long-standing questions about embryonic development.
In essence, researchers can zoom in on a tissue’s pointillist parts.
Sloan Kettering Institute Developmental Biology Program Chair Kat Hadjantonakis is keen to exploit the power of single-cell analysis. She wants to use the method to answer long-standing questions about how we develop as embryos. One in particular has stoked her passion: What happens to cells in extraembryonic tissues, such as the placenta, as an embryo grows?
“The dogma in the field of mammalian embryonic development was that extraembryonic tissues, such as the placenta, are just that — they’re external to the embryo,” she says. “They support the embryo during its in utero development, but they’re dispensable for adult life because the second you are born, you shed your extraembryonic tissue.”
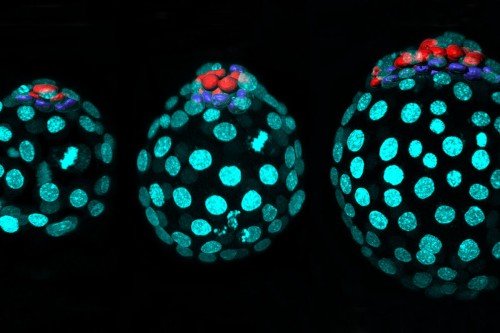
But what Dr. Hadjantonakis and her co-investigators have found with the help of single-cell analysis is that, in mice, cells from extraembryonic sources and cells from the embryo do mix. Adult tissues, such as the gut, in fact, contain mixtures of the two. The results alter the story of mammalian development. They also raise the question of how these extraembryonic interlopers might impact cancer.
Decisions, Decisions
Using cutting-edge microscopic imaging methods, Dr. Hadjantonakis’s team first observed that mouse embryos contained cells from extraembryonic tissues more than a decade ago. At that time, the discovery elicited more questions than answers. One question has persisted ever since: “Are these cells different based on where they come from?” she asks. “We could never really address that question because a technique to do so didn’t exist.”
Now, with single-cell analysis, it does. Single-cell analysis had to await the development of computational methods powerful enough to handle the massive amount of complex data that comes from sequencing the gene products of thousands of individual cells at once. To wield these methods appropriately, training in computational biology is required.

Computational biologist Dana Pe’er is an expert in single-cell RNA sequencing.
That’s why Dr. Hadjantonakis teamed up with SKI computational biologist Dana Pe’er, a world-renowned expert in single-cell analysis who also possesses the mathematic acumen needed to interpret this kind of data. They used the technique to analyze gene activity in nearly 120,000 individual cells of the developing mouse embryo and published their findings in the journal Nature in April 2019.
From these data, they were able to show, beyond a doubt, that mouse embryos contain extraembryonic cells. The cells could be distinguished on the basis of their gene activity.
“The idea that the major internal organs in our body are a mosaic mixture of embryonic and extraembryonic tissues was shocking to me,” says Dr. Pe’er, who is Chair of the Computational and Systems Biology Program at SKI. “It’s even more surprising than if you told me that the heart was made up of both heart cells and cells from the brain.”
Whether this patchwork of cells with different origins matters for the functioning of major internal organs is unknown. The SKI researchers think that the differences may not count as much when tissue is healthy but could have consequences during disease.

The Pe’er lab uses powerful computational methods to analyze vast amounts of data, with the goal of answering fundamental questions about biomedical science.
“One of the things that is being discovered — in my lab and in others — is that cancer cells tend to revert to developmentally more primitive states to do their evil deeds,” Dr. Pe’er says.
“Some of the nastiest cancers are those involving internal organs, like the lungs, gut, pancreas, and liver. For combating them, understanding the normal development of these tissues is going to be critical,” she adds.
In biology, as in art, getting up close and personal with the subject can change the way you see the world.